Difference between BERT and CharacterBERT
CharacterBERT is a variant of BERT that tries to go back to the simpler days where models produced single embeddings for single words (or rather, tokens). In practice, the only difference is that instead of relying on WordPieces, CharacterBERT uses a CharacterCNN module just like the one that was used in ELMo [1].
The next figure show the inner mechanics of the CharacterCNN and compares it to the original WordPiece system in BERT.
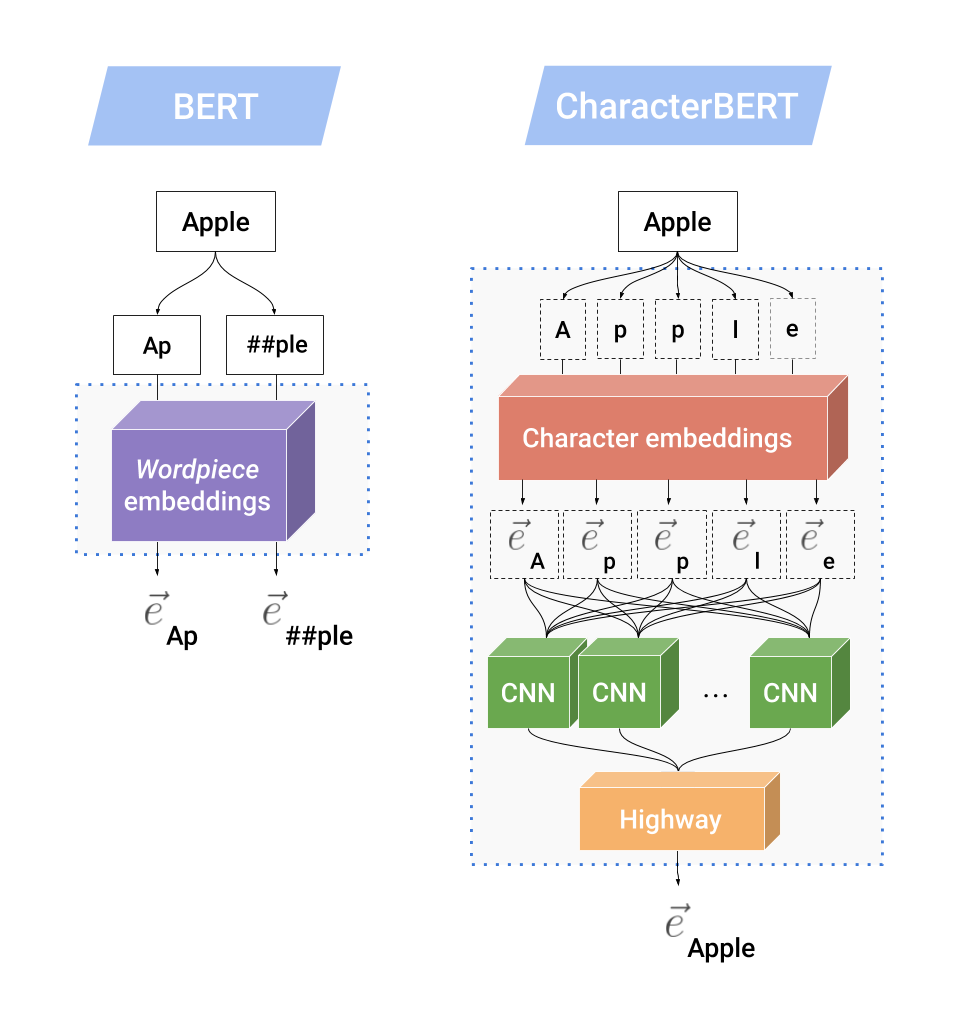
Let’s imagine that the word “Apple” is an unknown word (i.e. it does not appear in BERT’s WordPiece vocabulary), then BERT splits it into known WordPieces: [Ap, ##ple], where ## are used to designate WordPieces that are not at the beginning of a word. Then, each subword unit is embedded using a WordPiece embedding matrix, producing two output vectors.
On the other hand, CharacterBERT does not have a WordPiece vocabulary and can handle any input token as long as it is not unreasonably long (i.e. under 50 characters). Instead of splitting “Apple”, CharacterBERT reads it as a sequence of characters: [A, p, p, l, e]. Each character is then represented using a character embedding matrix, producing a sequence of character embeddings. This sequence is then fed to multiple CNNs, each responsible for scanning the sequence n-characters at a time, with n =[1..7]. All CNN outputs are aggregated into a single vector that is then projected down to the desired dimension using Highway Layers [3]. This final projection is the context-independent representation of the word “Apple”, which will be combined with position and segment embeddings before being fed to multiple Transformer Layers as in BERT.
Reasons for CharacterBERT instead of BERT
CharacterBERT acts almost as a drop in replacement for BERT that
- Produces a single embedding for any input token
- Does not rely on a WordPiece vocabulary
The first point is clearly desirable as working with single embeddings is far more convenient than having a variable number of WordPiece vectors for each token. As for the second point, it is particularly relevant when working in specialized domains (e.g. medical domain, legal domain, …). In fact, the common practice when building specialized versions of BERT (e.g. BioBERT [4], BlueBERT [5] and some SciBERT [6] models) is to re-train the original model on a set of specialized texts. As a result, most SOTA specialized models keep the original general-domain WordPiece vocabulary which is not suited for specialized domain applications.
The table below shows the difference between the original general-domain vocabulary and a medical WordPiece vocabulary that was built on medical corpora: MIMIC [7] and PMC OA [8].
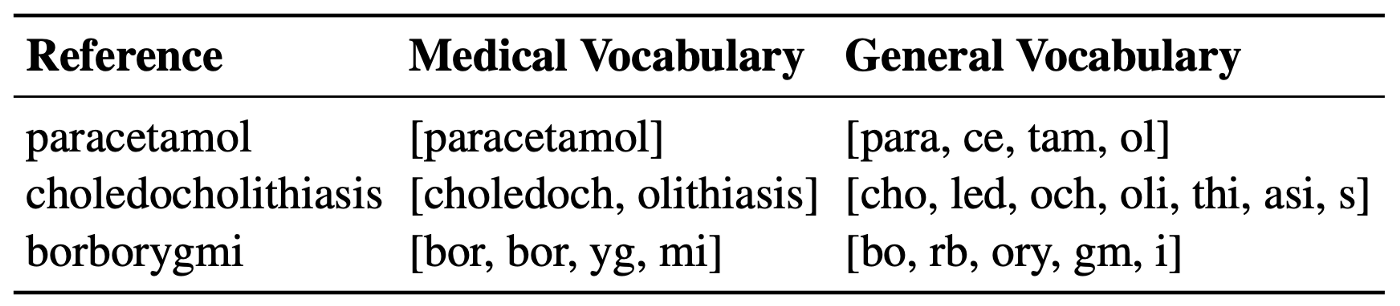
We can clearly see that BERT’s vocabulary is not very well suited for specialized terms (e.g. “choledocholithiasis” is split into [cho, led, och, oli, thi, asi, s]). The medical wordpiece works better however it has its limits as well (e.g. “borborygmi” into [bor, bor, yg, mi]). Therefore, in order to avoid any biases that may come from using the wrong WordPiece vocabulary, and in an effort to got back to conceptually simpler models, a variant of BERT was proposed: CharacterBERT.
BERT vs. CharacterBERT
BERT and CharacterBERT are compared in a classic scenario where a general model is first pre-trained before serving as an initialisation for the pre-training of a specialized version.
Note: we focus here on the English language and the medical domain.
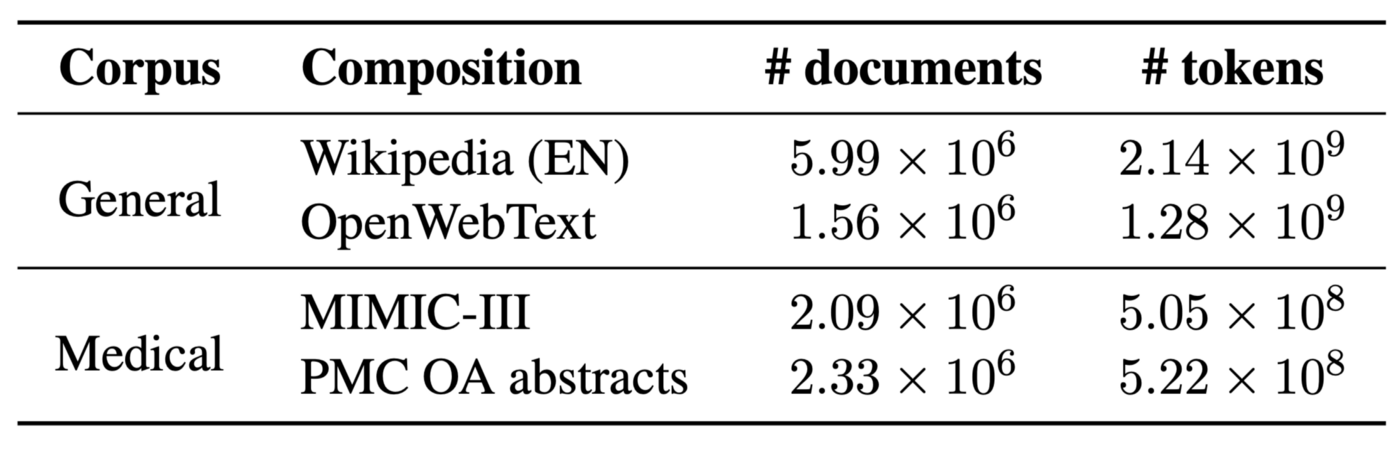
To be as fair as possible, both BERT and CharacterBERT are pre-trained from scratch in exactly the same conditions. Then, each pre-trained model is evaluated on multiple medical tasks. Let’s take an example.
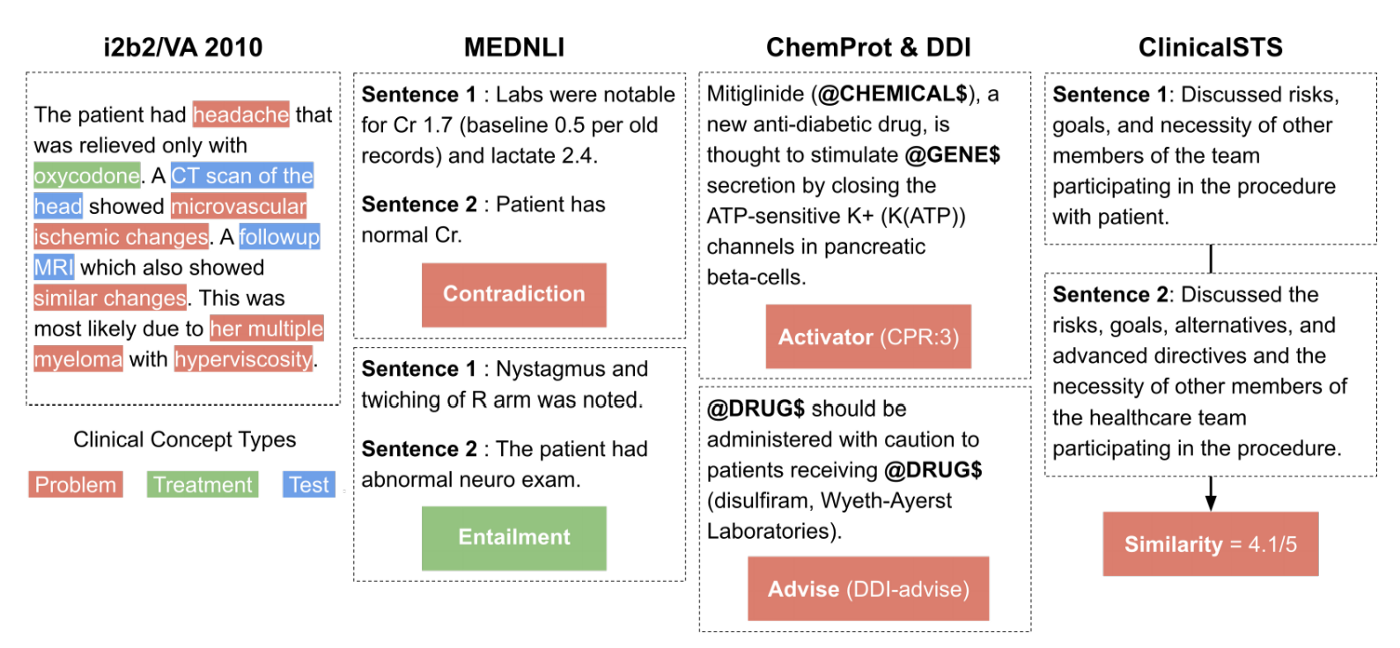
i2b2/VA 2010 [9] is a competition that consists in multiple tasks including the clinical concept detection task which was used to evaluate our models. The goal is to detect three types of clinical concepts: Problem, Treatment and Test. An example is given in the far left section of the figure above.
As usual, we evaluate our models by first training on the training set. At each iteration, the model is tested on a separate validation set, allowing us to save the best iteration. Finally, after going through all the iterations, a score (here a strict F1 score) is computed on the test set using the model from the best iteration. This whole procedure is then repeated 9 more times using different random seeds, which allows us to account for some of the variance and report final model performances as: mean ± std.
Note: more details are available in the paper [2]
Evaluation Results
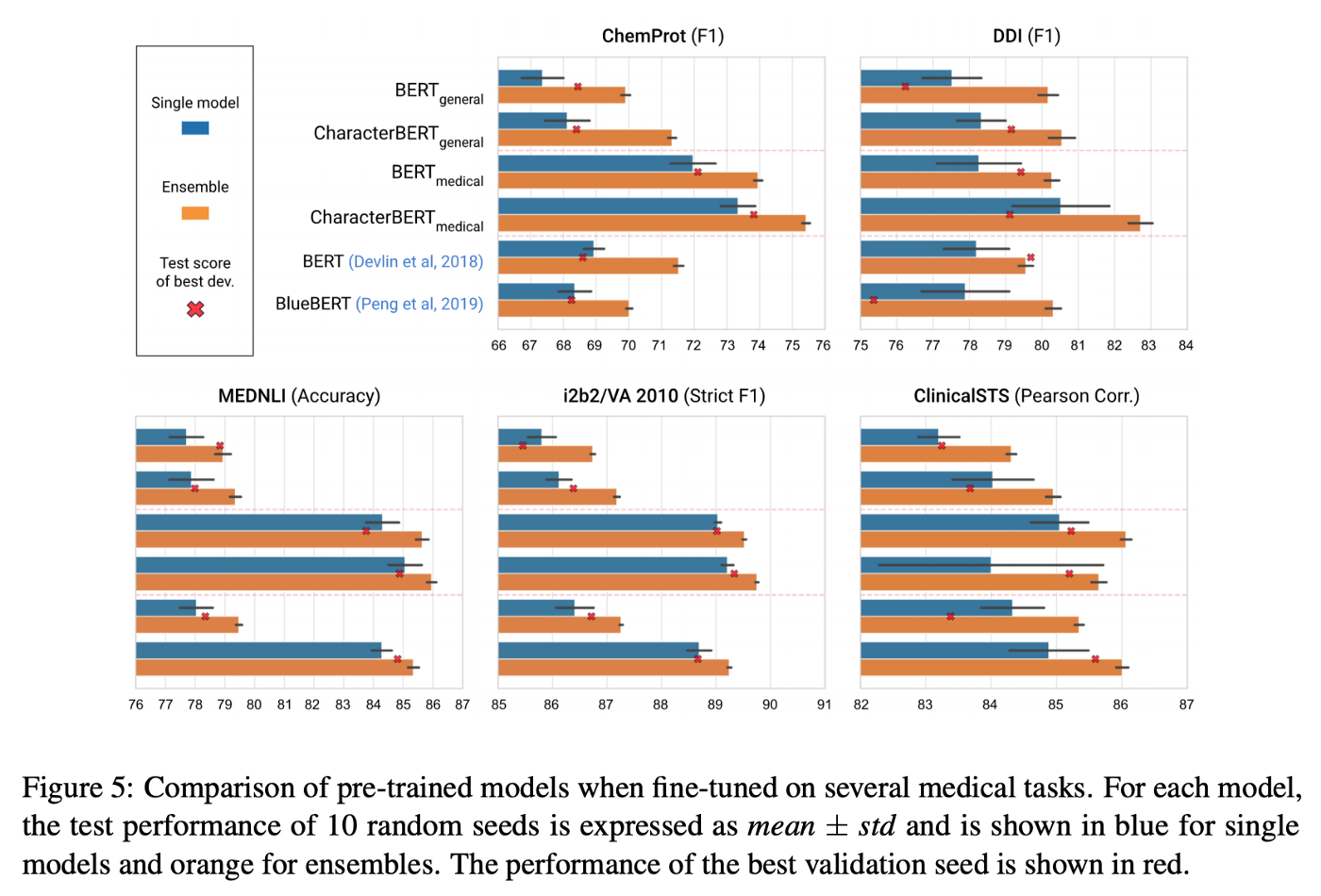
In most cases, CharacterBERT outperformed its BERT counterpart.
Note: The only exception is the ClinicalSTS task where the medical CharacterBERT got (on average) a lower score than the BERT version. This may be due to the task dataset being small (1000 examples vs. 30,000 on average for other tasks) and should be investigated.
Robustness to Noise
Besides pure performance, another interesting aspect is whether the models are robust to noisy inputs. In fact, we evaluated BERT and CharacterBERT on noisy versions of the MedNLI task [10] where (put simply) the goal is to say whether two medical sentences are in contradiction with each other. Here, a noise level of X% means that each character in the text if either replaced or swapped with a X% probability. The results are displayed on the figure below.
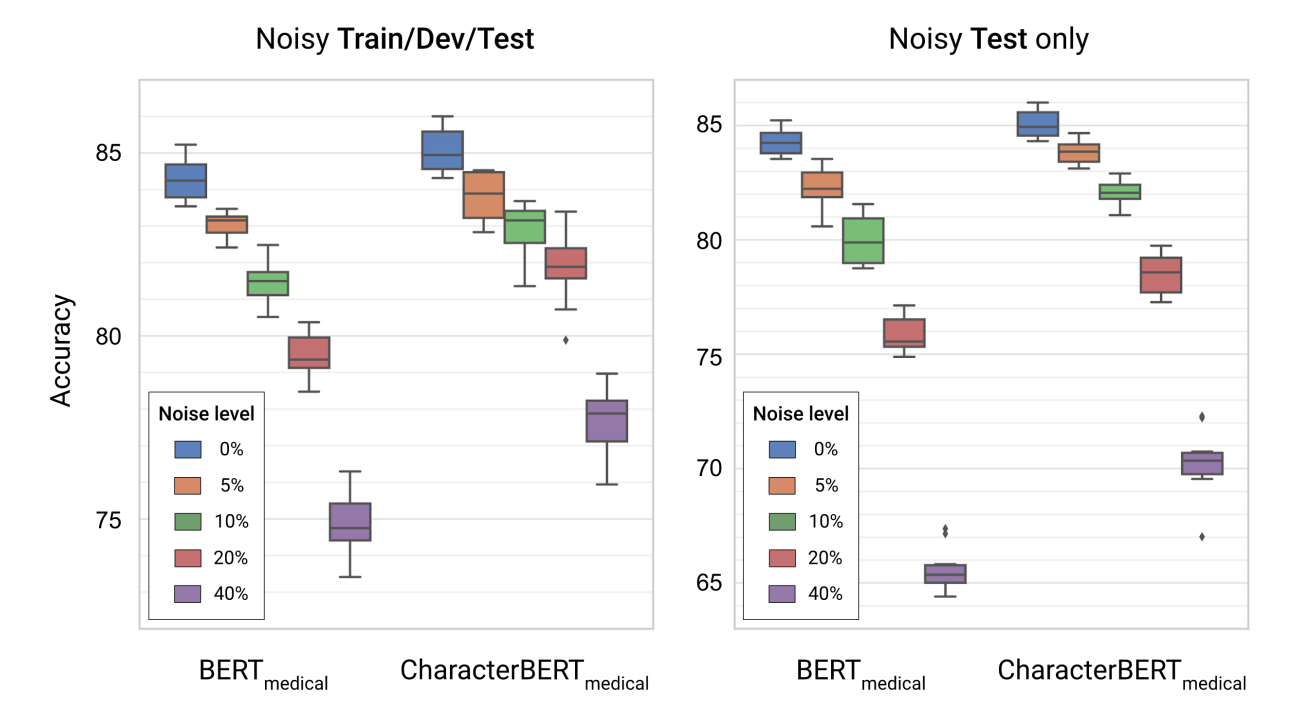
As you can see, the medical CharacterBERT model seems to be more robust than medical BERT: the initial gap between the two models of ~1% accuracy grows to ~3% when adding noise to all splits, and ~5% when surprising the models with noise only in the test set.
Downsides of CharacterBERT
The main downside of CharacterBERT is its slower pre-training speed. This is due to:
- the CharacterCNN module that is slower to train;
- but mainly because the model is working at the token-level:
it updates a large token vocabulary at each pre-training iteration.
Note: However, CharacterBERT is just as fast as BERT during inference (actually, it is even a bit faster) and pre-trained models are available so you can skip the pre-training step altogether 😊!
Conclusion
All in all, CharacterBERT is a simple variant of BERT that replaces the WordPiece system with a CharacterCNN (just like ELMo before that). Evaluation results on multiple medical tasks show that this change is beneficial: improved performance & improved robustness to misspellings. Hopefully, this model will motivate more research towards word-level open-vocabulary transformer-based language models (e.g. applying the same idea to ALBERT [11], ERNIE [12]).
Original paper:
https://arxiv.org/abs/2010.10392
Code & pre-trained models:
https://github.com/helboukkouri/character-bert
Presentation Slides:
CharacterBERT.pdf
Work done by:
Hicham El Boukkouri (myself), Olivier Ferret, Thomas Lavergne, Hiroshi Noji, Pierre Zweigenbaum and Junichi Tsujii
References
- [1] Peters, Matthew E., et al. “Deep contextualized word representations.” arXiv preprint arXiv:1802.05365 (2018).
- [2] El Boukkouri, Hicham, et al. “CharacterBERT: Reconciling ELMo and BERT for Word-Level Open-Vocabulary Representations From Characters.” arXiv preprint arXiv:2010.10392 (2020).
- [3] Srivastava, Rupesh Kumar, Klaus Greff, and Jürgen Schmidhuber. “Highway networks.” arXiv preprint arXiv:1505.00387 (2015).
- [4] Lee, Jinhyuk, et al. “BioBERT: a pre-trained biomedical language representation model for biomedical text mining.” Bioinformatics 36.4 (2020): 1234–1240.
- [5] Peng, Yifan, Shankai Yan, and Zhiyong Lu. “Transfer learning in biomedical natural language processing: An evaluation of bert and elmo on ten benchmarking datasets.” arXiv preprint arXiv:1906.05474 (2019).
- [6] Beltagy, Iz, Kyle Lo, and Arman Cohan. “SciBERT: A pretrained language model for scientific text.” arXiv preprint arXiv:1903.10676 (2019).
- [7] Johnson, Alistair, et al. “MIMIC-III Clinical Database” (version 1.4). PhysioNet (2016), https://doi.org/10.13026/C2XW26.
- [8] PMC OA corpus: https://www.ncbi.nlm.nih.gov/pmc/tools/openftlist/
- [9] Uzuner, Özlem, et al. “2010 i2b2/VA challenge on concepts, assertions, and relations in clinical text.” Journal of the American Medical Informatics Association 18.5 (2011): 552–556.
- [10] Shivade, Chaitanya. “MedNLI - A Natural Language Inference Dataset For The Clinical Domain” (version 1.0.0). PhysioNet (2019), https://doi.org/10.13026/C2RS98.
- [11] Lan, Zhenzhong, et al. “ALBERT: A lite BERT for self-supervised learning of language representations.” arXiv preprint arXiv:1909.11942 (2019).
- [12] Sun, Yu, et al. “ERNIE 2.0: A Continual Pre-Training Framework for Language Understanding.” AAAI. 2020.